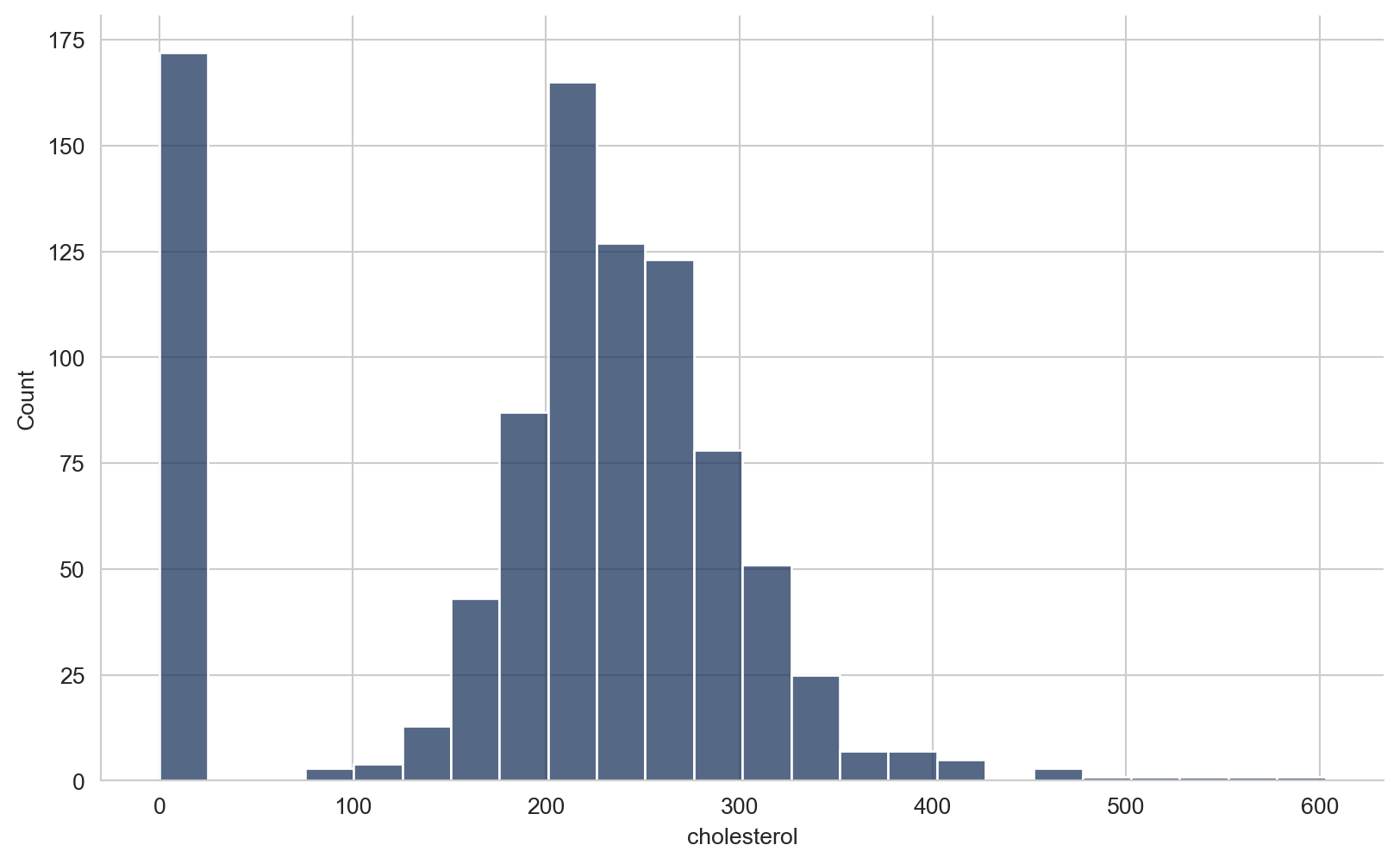
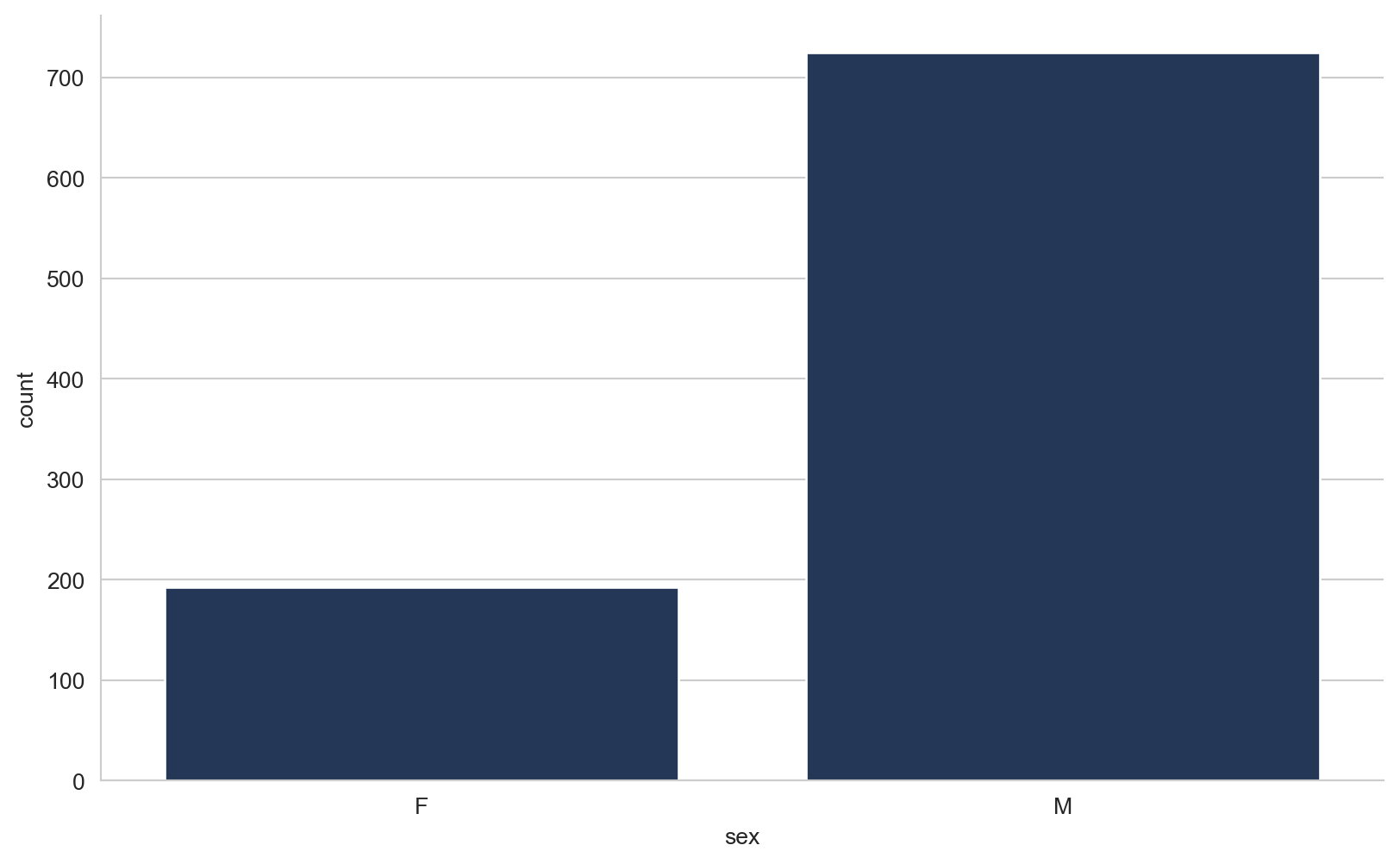
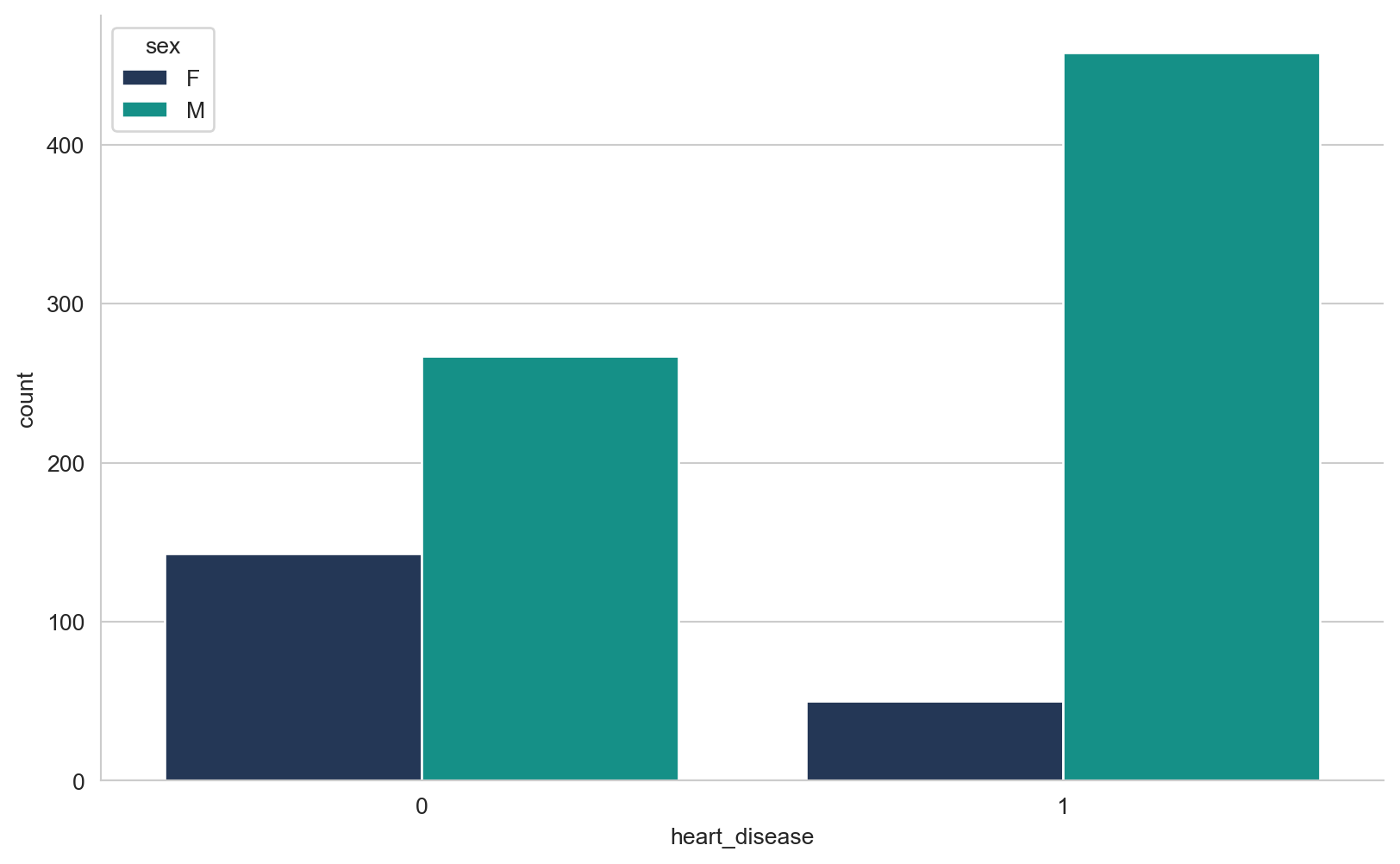
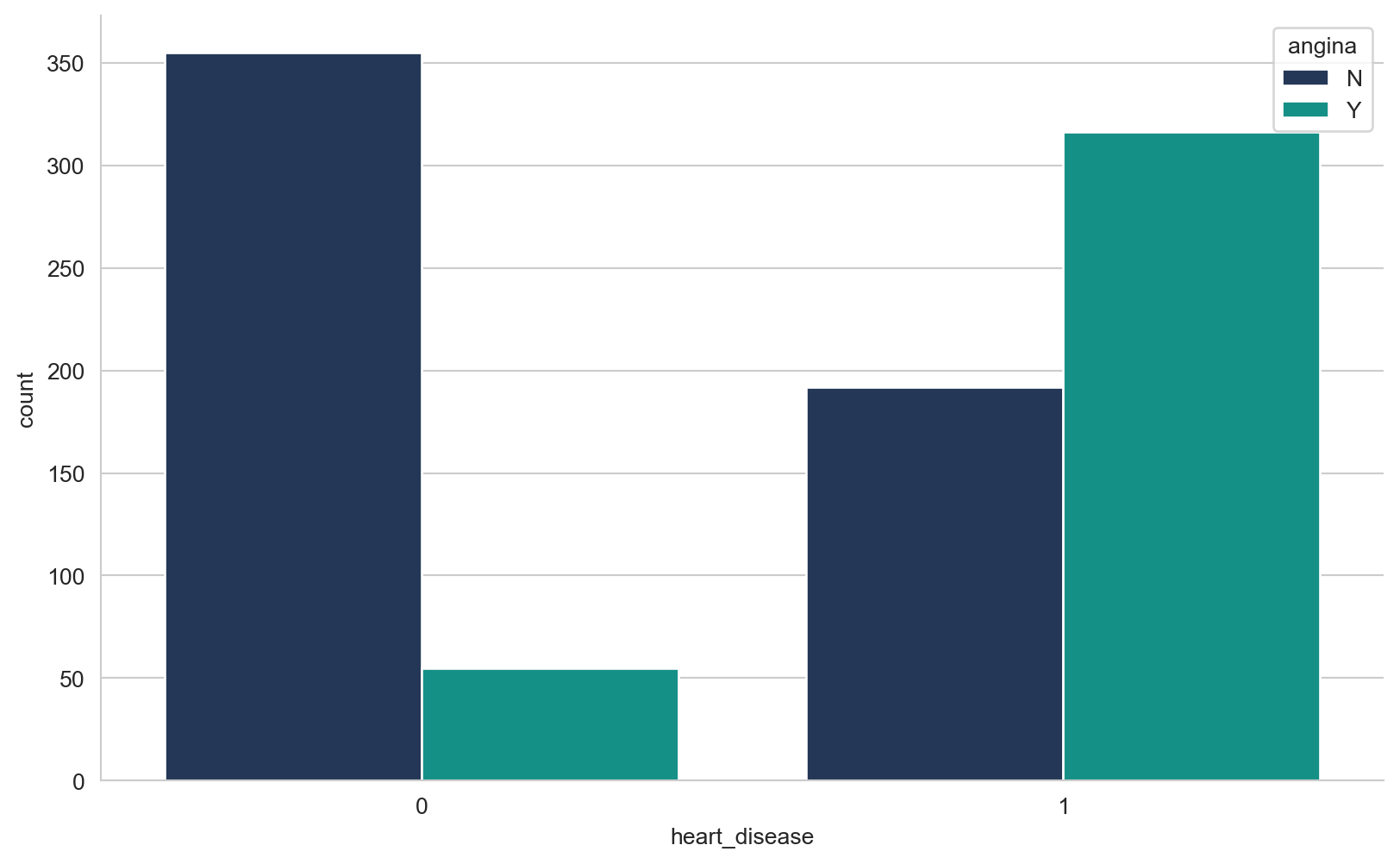
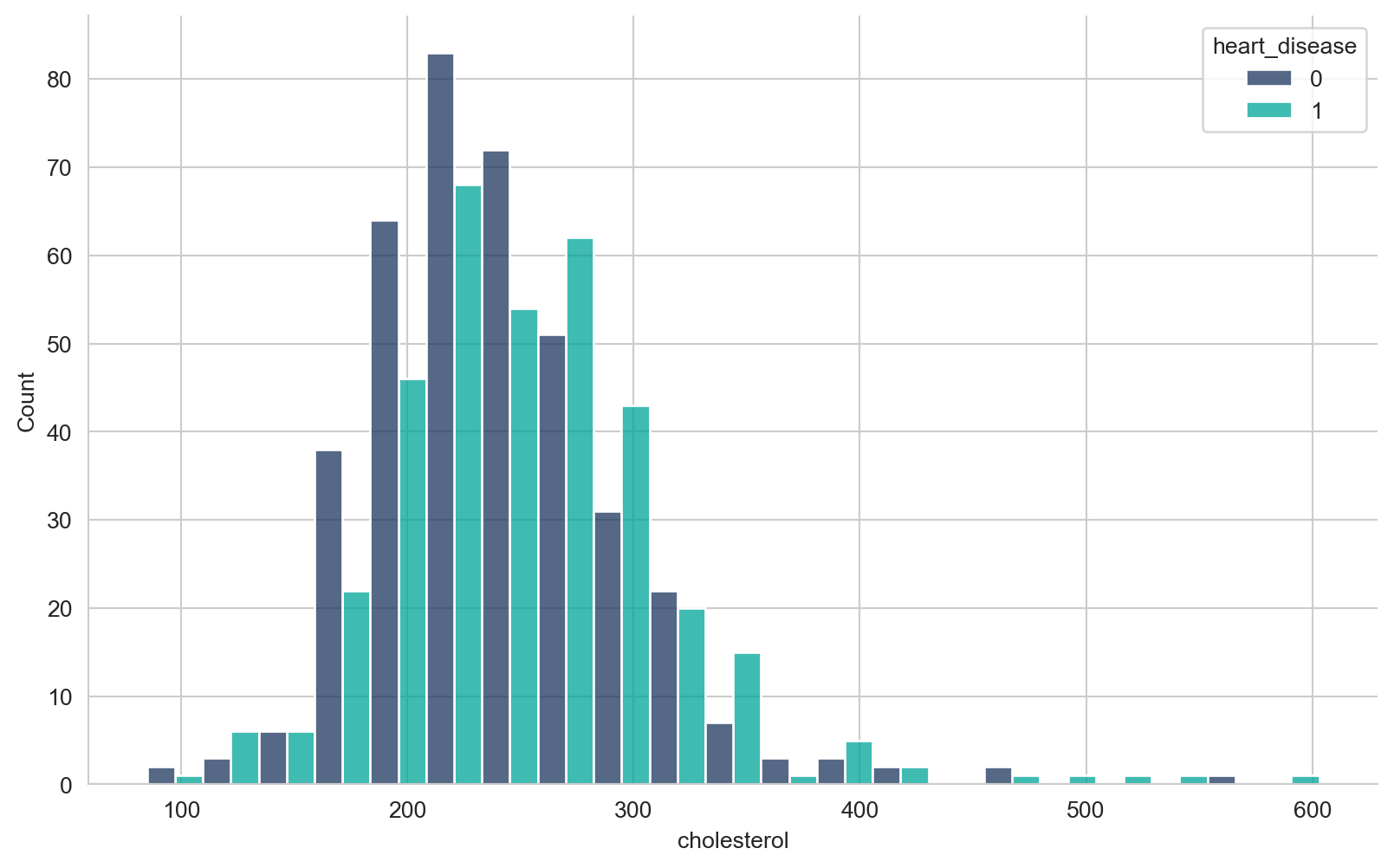
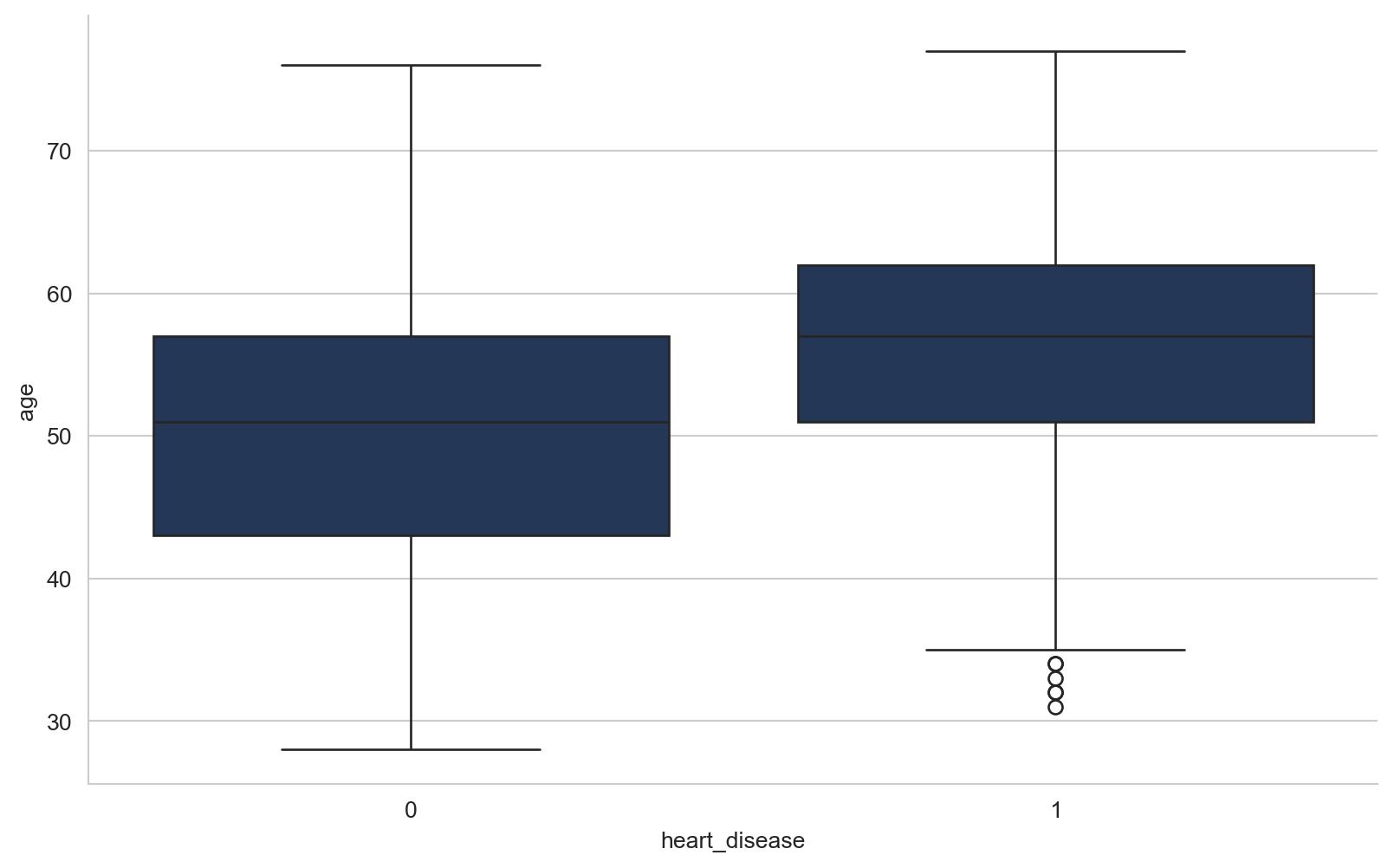
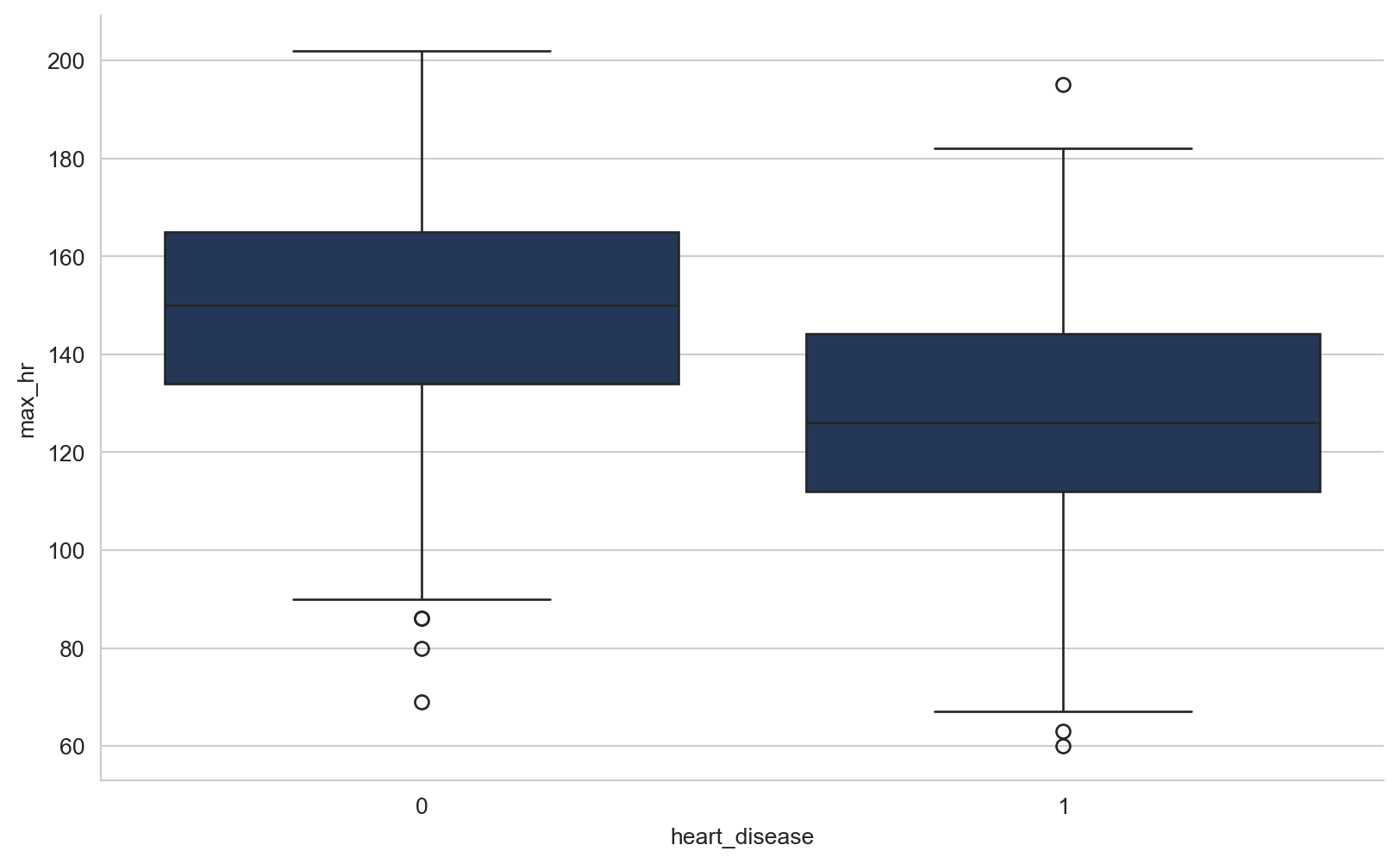
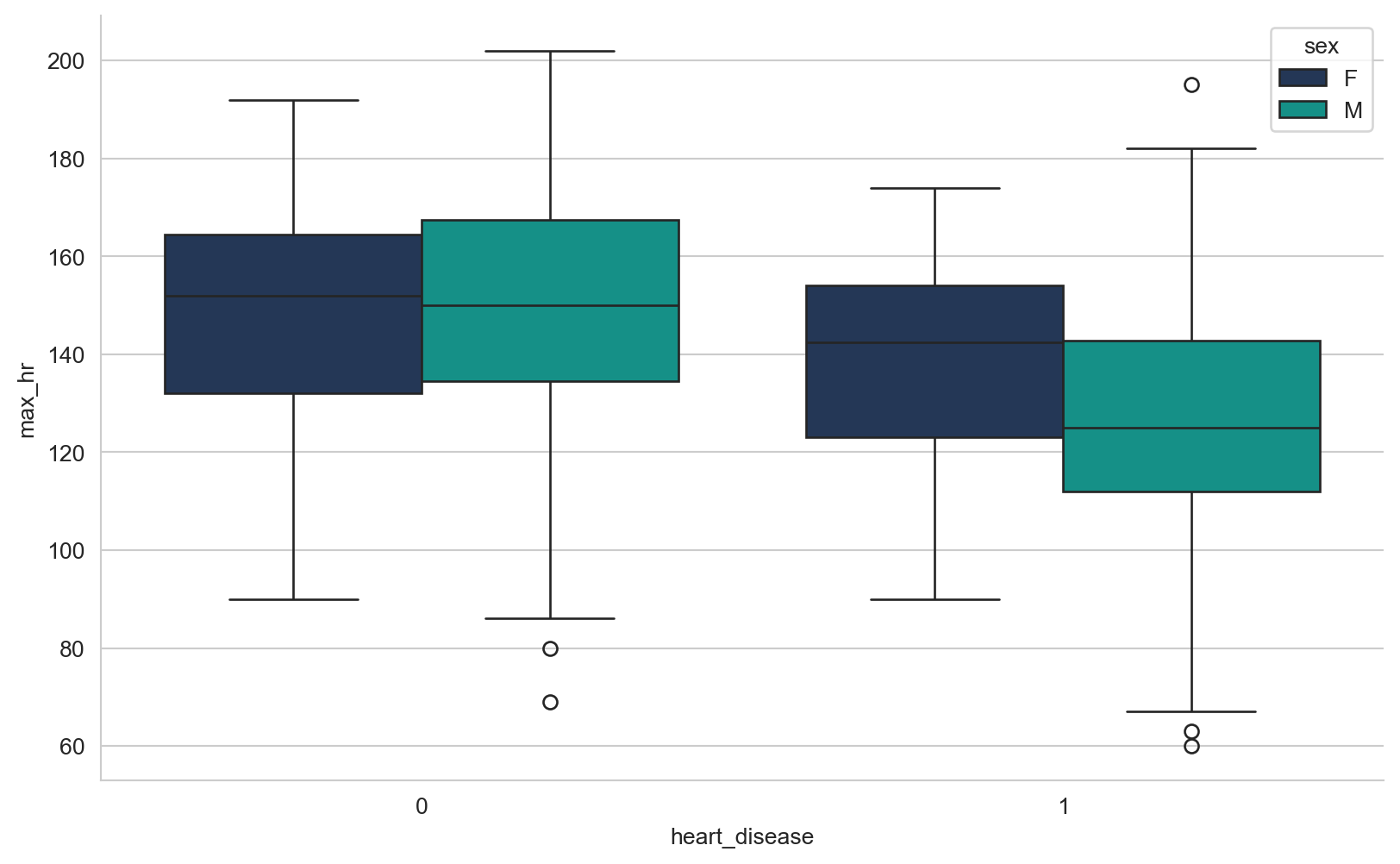
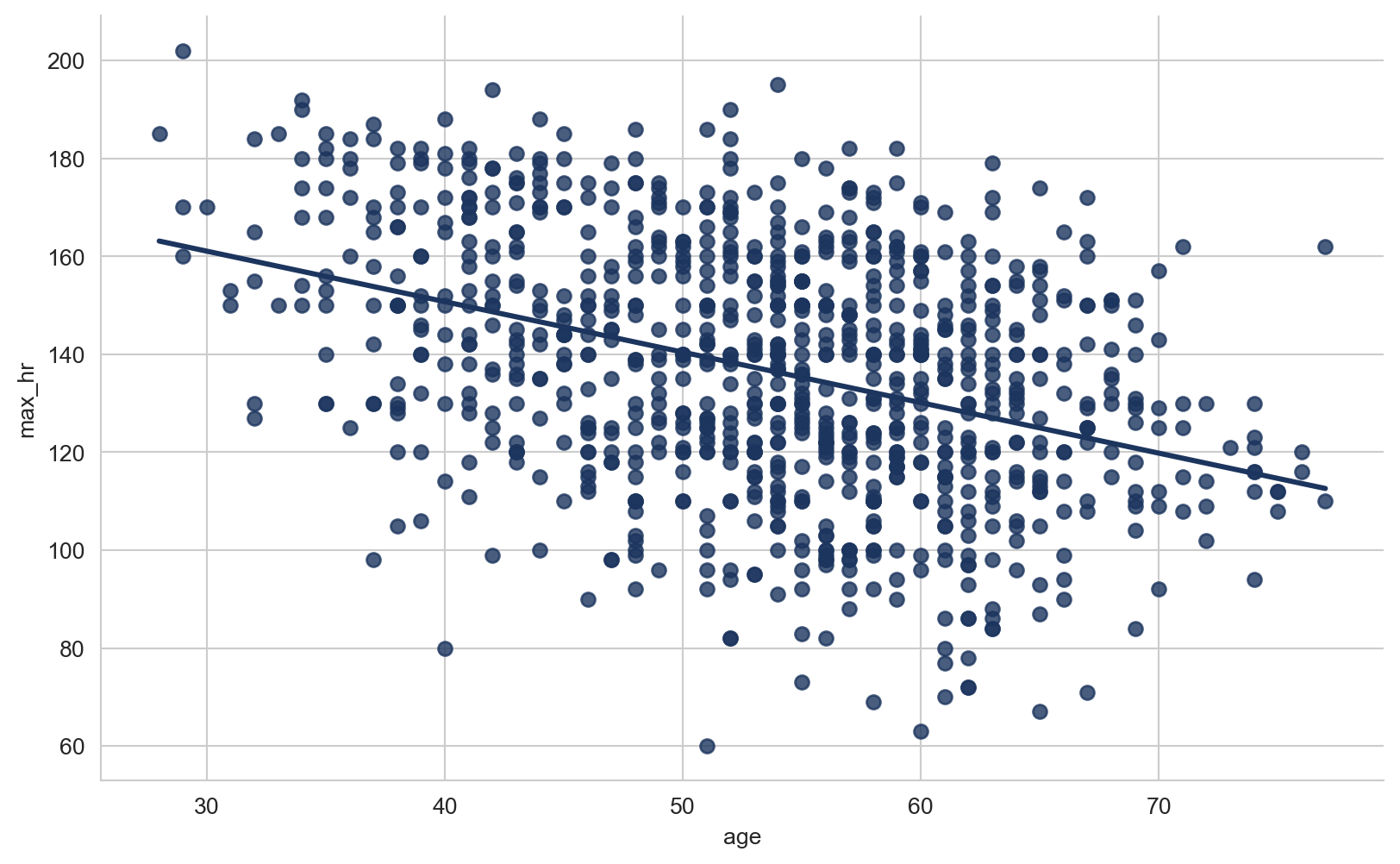
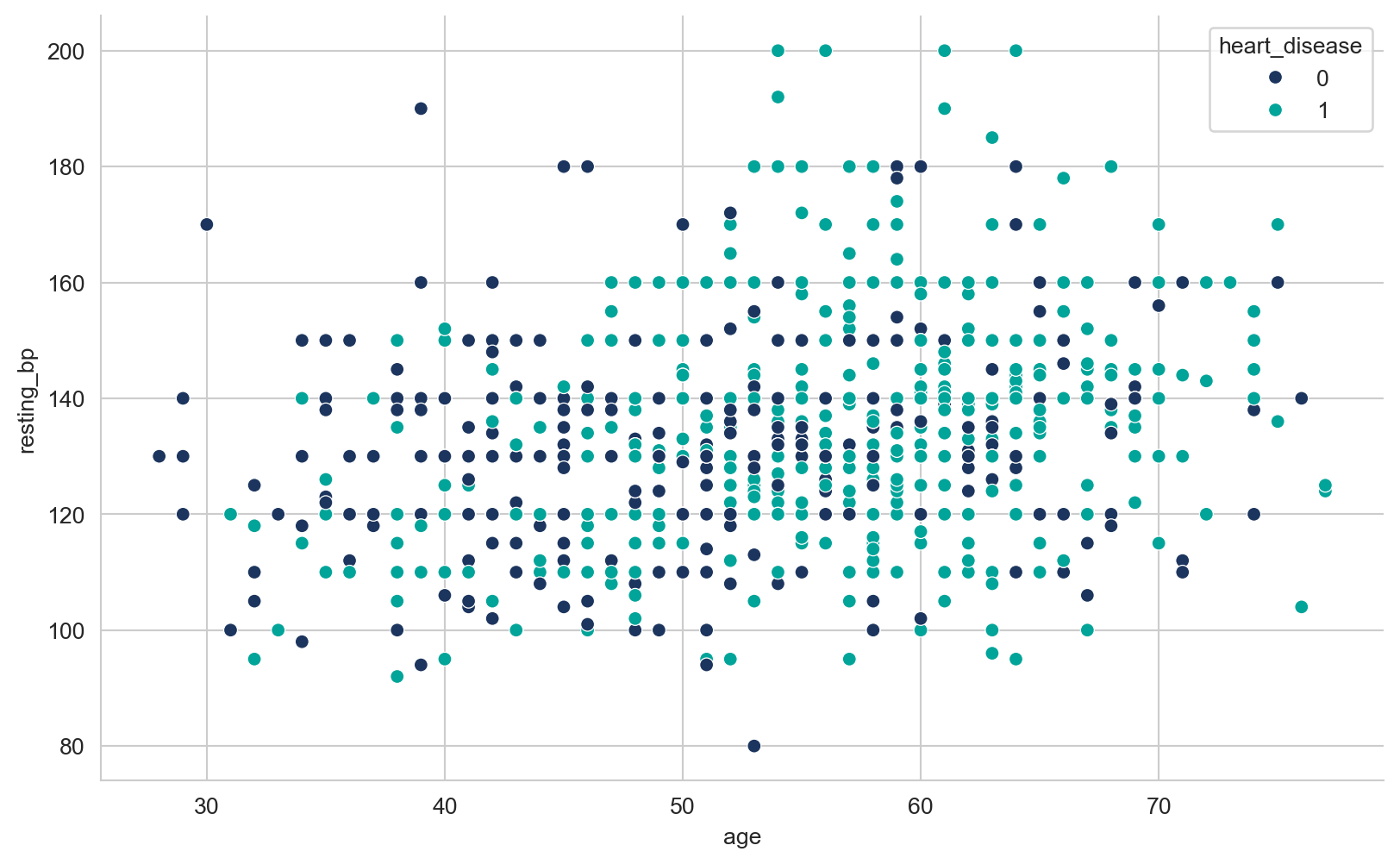
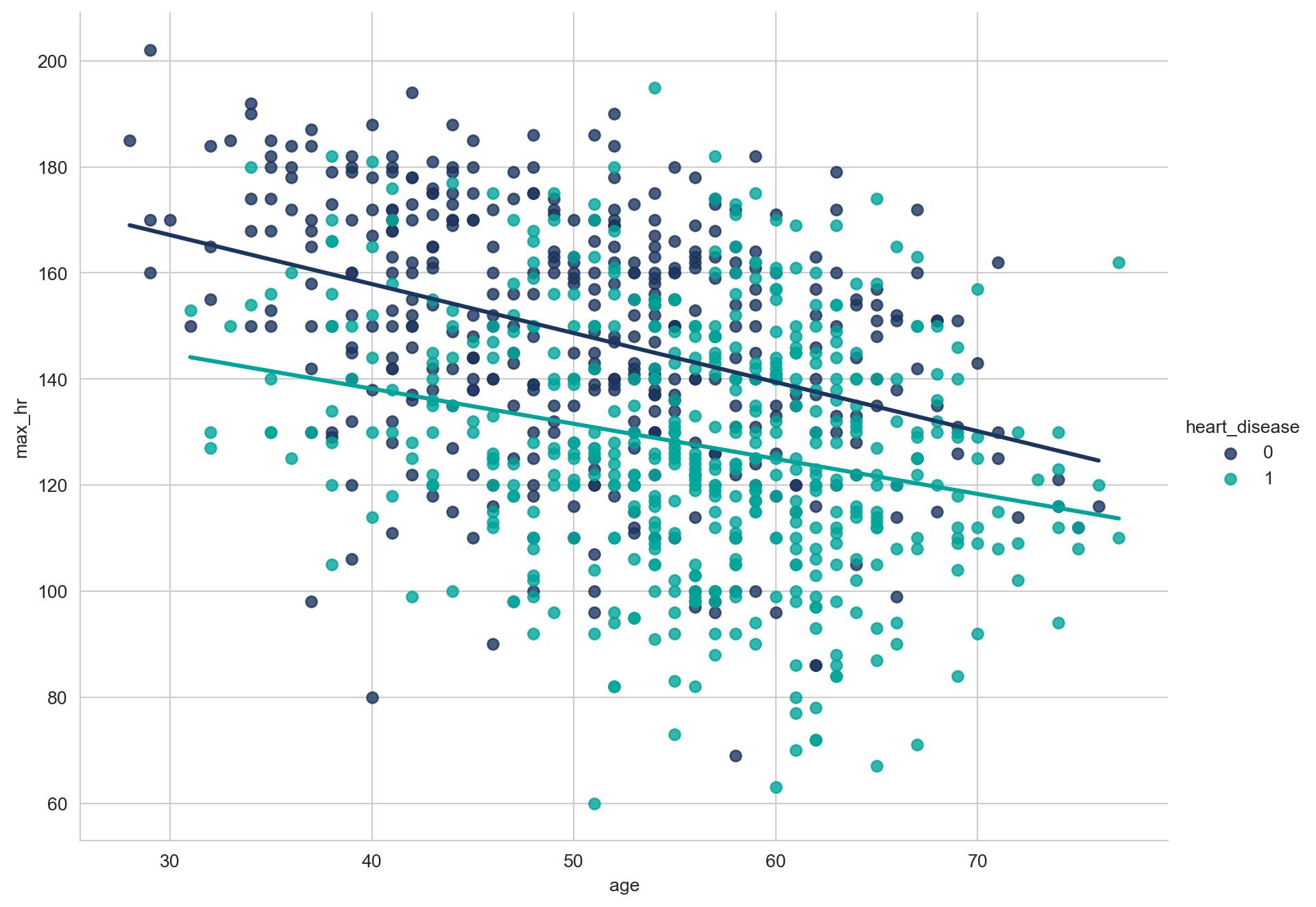
array([289, 180, 283, 214, 195, 339, 237, 208, 207, 284, 211, 164, 204,
234, 273, 196, 201, 248, 267, 223, 184, 288, 215, 209, 260, 468,
188, 518, 167, 224, 172, 186, 254, 306, 250, 177, 227, 230, 294,
264, 259, 175, 318, 216, 340, 233, 205, 245, 194, 270, 213, 365,
342, 253, 277, 202, 297, 225, 246, 412, 265, 182, 218, 268, 163,
529, 100, 206, 238, 139, 263, 291, 229, 307, 210, 329, 147, 85,
269, 275, 179, 392, 466, 129, 241, 255, 276, 282, 338, 160, 156,
272, 240, 393, 161, 228, 292, 388, 166, 247, 331, 341, 243, 279,
198, 249, 168, 603, 159, 190, 185, 290, 212, 231, 222, 235, 320,
187, 266, 287, 404, 312, 251, 328, 285, 280, 192, 193, 308, 219,
257, 132, 226, 217, 303, 298, 256, 117, 295, 173, 315, 281, 309,
200, 336, 355, 326, 171, 491, 271, 274, 394, 221, 126, 305, 220,
242, 347, 344, 358, 169, 181, 0, 236, 203, 153, 316, 311, 252,
458, 384, 258, 349, 142, 197, 113, 261, 310, 232, 110, 123, 170,
369, 152, 244, 165, 337, 300, 333, 385, 322, 564, 239, 293, 407,
149, 199, 417, 178, 319, 354, 330, 302, 313, 141, 327, 304, 286,
360, 262, 325, 299, 409, 174, 183, 321, 353, 335, 278, 157, 176,
131], dtype=int64)